Acknowledging NSF for their support: “Funded by the US National Science Foundation, DBI-1922642”.
Publications
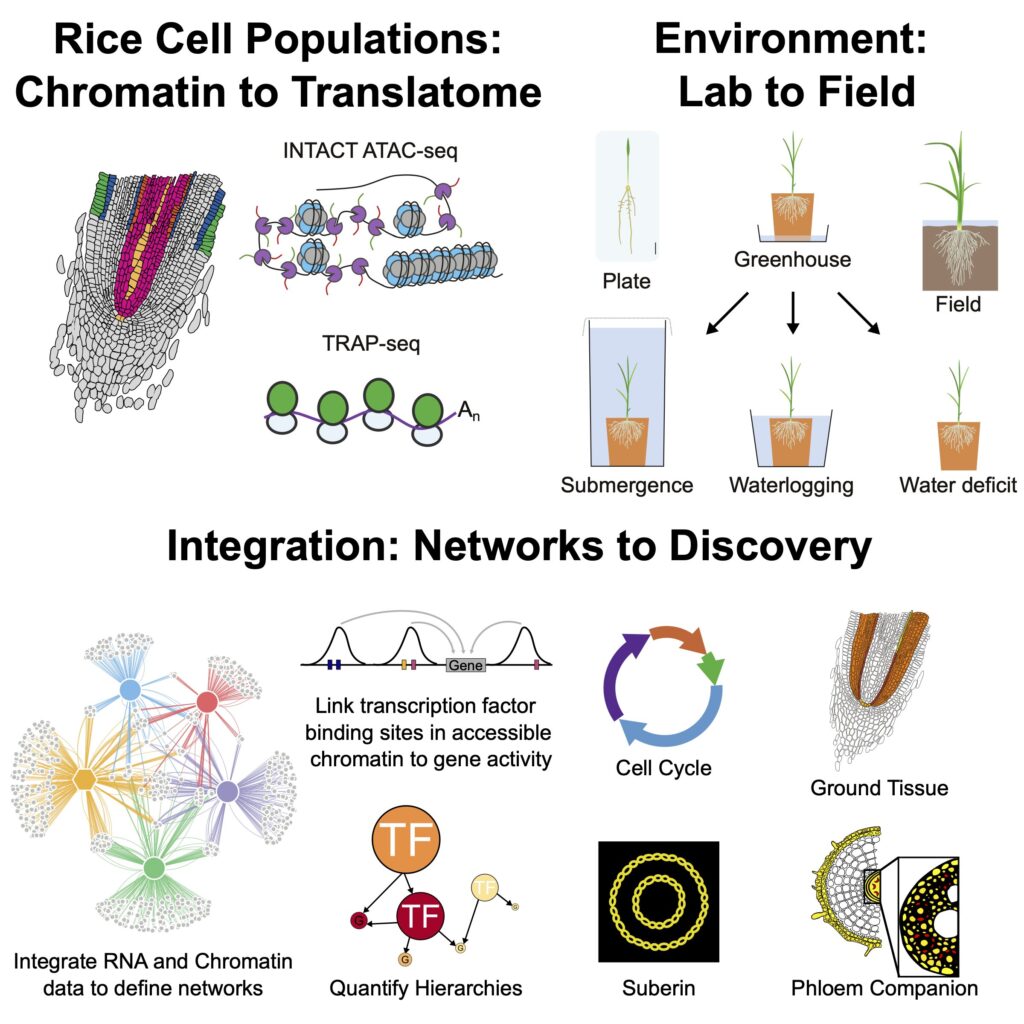
- Reyonoso et al., 2022. Gene regulatory networks shape developmental plasticity of root cell types under water extremes in rice. https://doi.org/10.1016/j.devcel.2022.04.013 Plants3D trainee Alex Borowsky is a co-author and contributer. Summary: In this paper, we provide an atlas of gene activity of root cell populations of rice in varied agronomically relevant environments. We profiled translatomes and accessible chromatin regions, which enabled us to define gene regulatory networks involved in processes including the cell cycle and suberin biosynthesis. In all, we identify genomic cis- and transregulatory factors involved in stress responses that can facilitate breeding and engineering climate-resilient crops.
- White et al., 2022. Rapid analysis of strigolactone receptor activity in a Nicotiana benthamiana dwarf14 mutant. https://onlinelibrary.wiley.com/doi/10.1002/pld3.389 Plants3D trainee Alex White is a co-author and contributer. Summary: In my research I created a mutant line of Nicotiana benthamiana. Specifically, I knocked out the receptor, D14, for strigolactone perception. Using this mutant, I developed a transient assay that allows us to observe how strigolactone dependent protein targets are degraded when co-expressed with different variants of D14 without interference from endogenous NbD14.
- Kajala et al., 2021. Cell; Innovation, conservation, and repurposing of gene function in root cell type development https://doi.org/10.1016/j.cell.2021.04.024. A UCR news release about this paper can be found here https://news.ucr.edu/articles/2021/05/18/discovery-increases-likelihood-growing-food-despite-drought. Plants3D trainee Alex Borowsky is a co-author and contributed to the phylogenetic analysis and scientific insight. Summary: Translatome profiles for different cell types in the roots of tomato were generated to show how different cell layers of roots, such as the exodermis and xylem, are defined at a molecular level. Additionally, a comparison of tomato, rice, and previously published Arabidopsis data meristematic stem cell regions of roots show greater cross-species conservation in gene expression than more differentiated, specialized cells. Alex contributed to the phylogenetic analyses needed to interpret the cross-species comparisons.
Posters
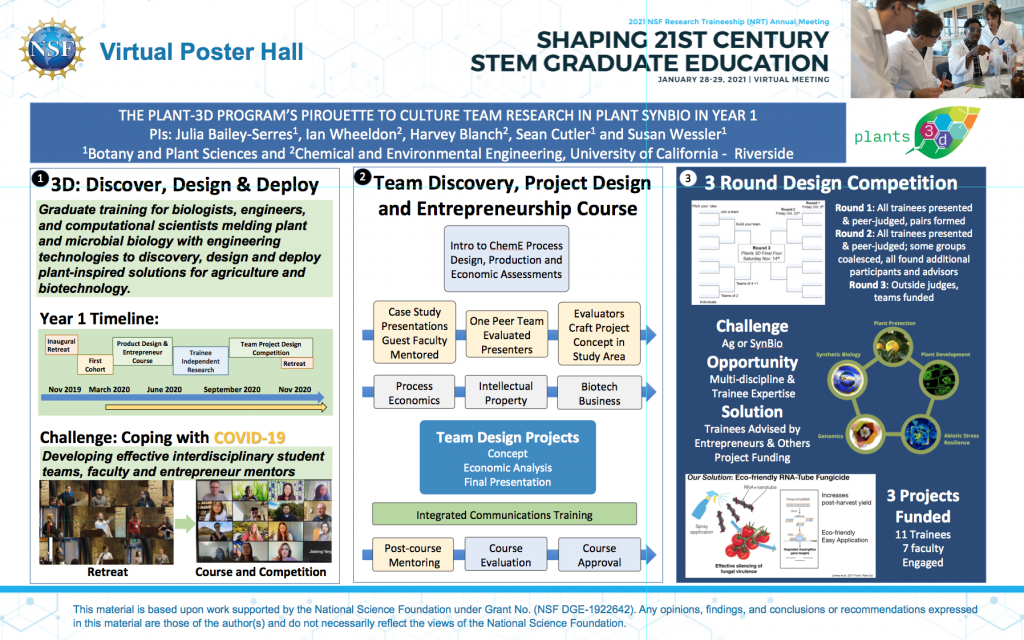
- NSF NRT PI Meeting 2021 Poster – Our Poster summarizing how we developed our program in year 1 with our amazingly adaptable trainees despite the challenges of COVID-19.
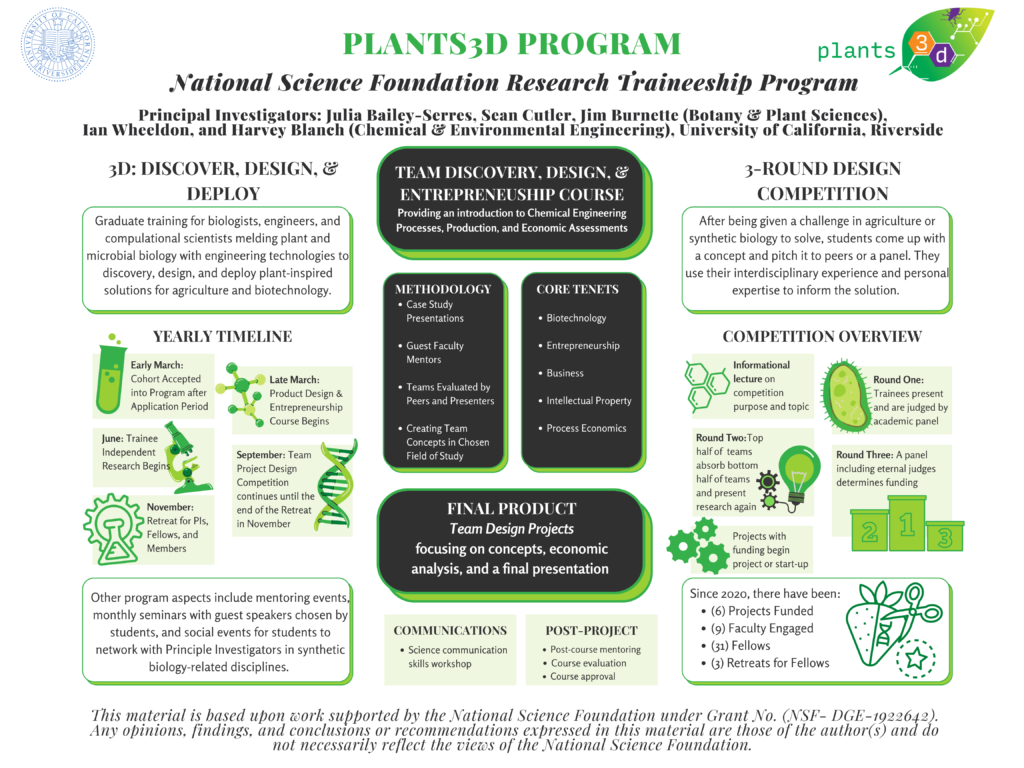
- Graduate Orientation Poster – A poster designed to display updates to the Plants3D Program.